Genetic architecture of nonhost resistance in grasses to fungal pathogens
Collaborations: Michael Ayliffe at CSIRO, Black Mountain, Wolfgang Spielmeyer at CSIRO, Black Mountain, Zakkie Pretorius University of the Free State, and Melania Figueroa at University of Minnesota
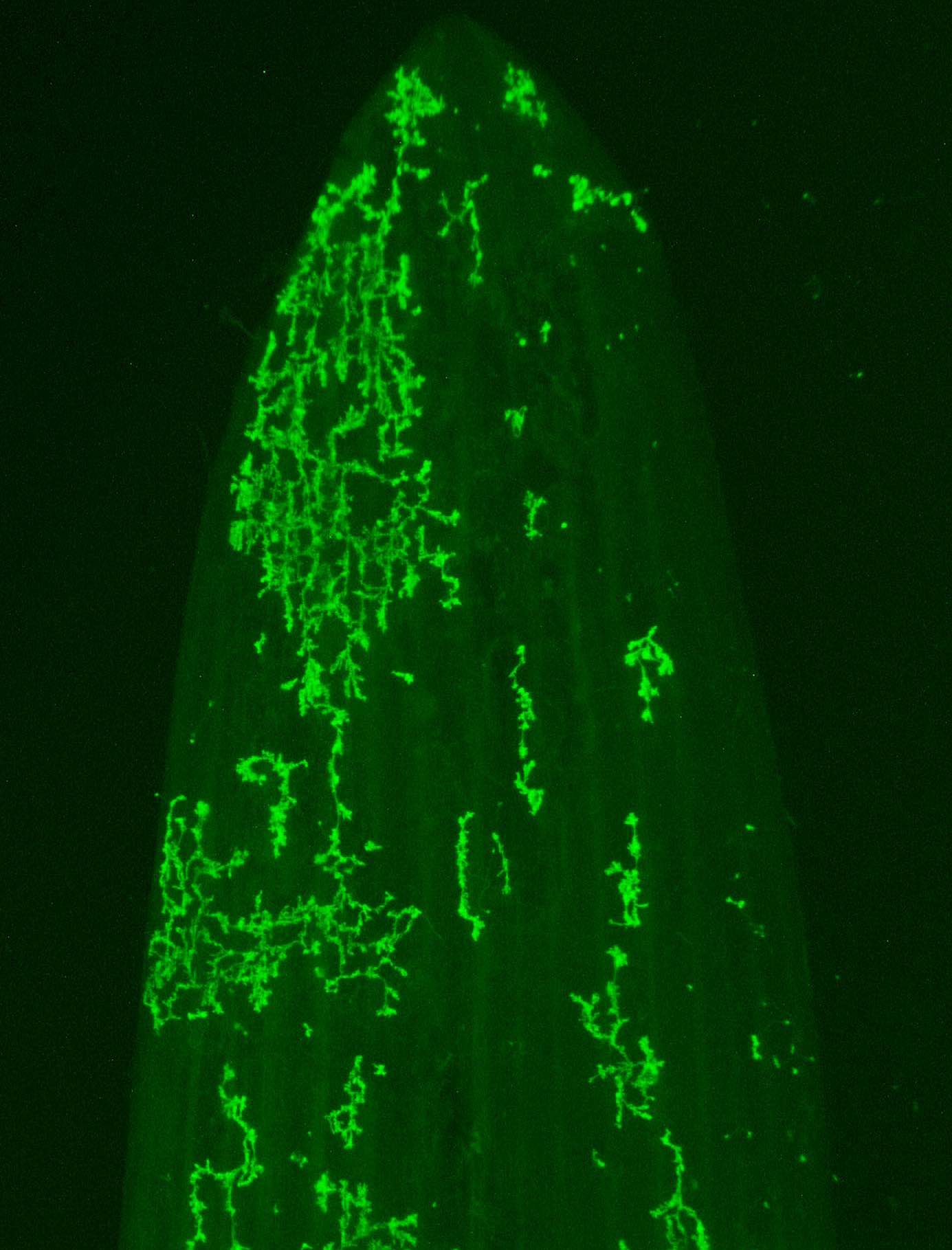
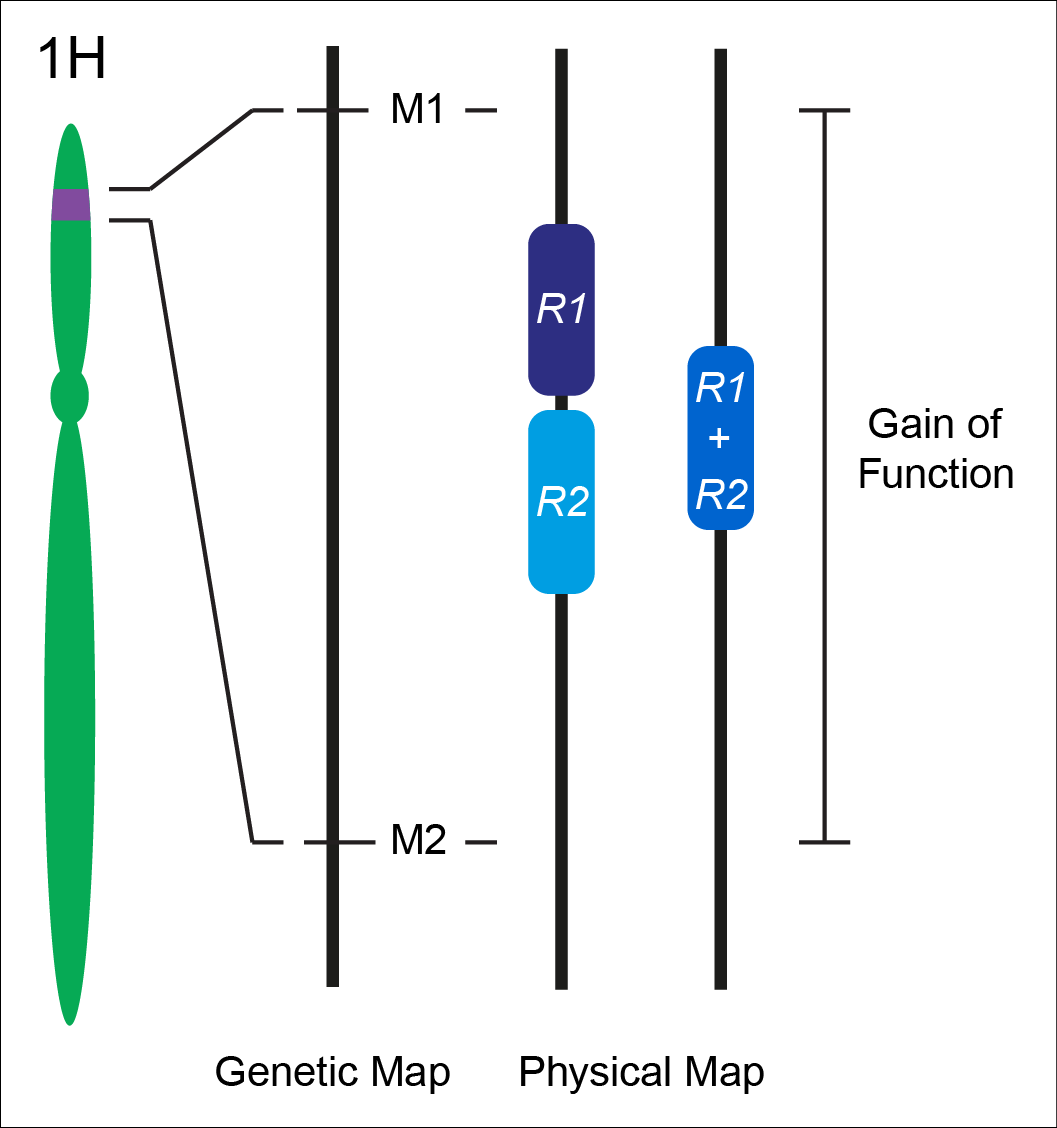
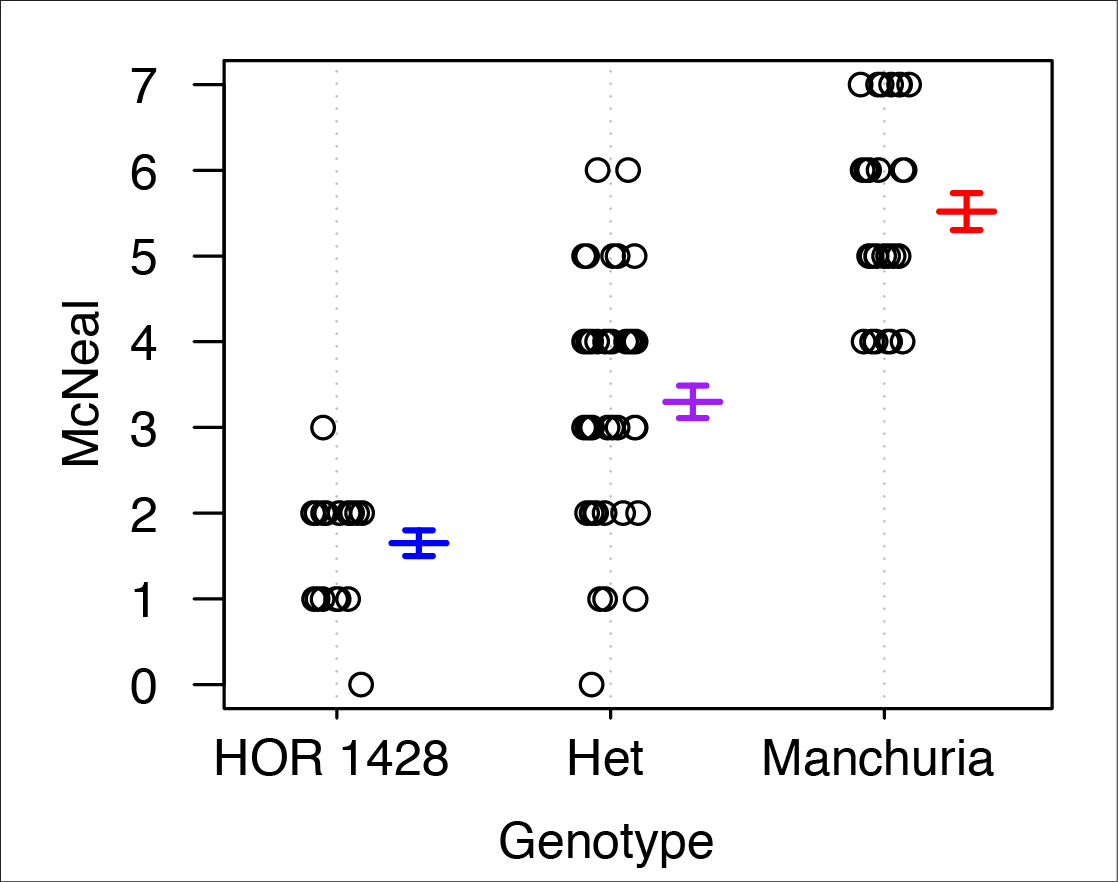
Nonhost resistance has long been considered a source for engineering durable disease resistance. While we have an extensive mechanistic understanding of the genes underlying host resistance, little is known about the genes conditioning resistance in the transition from host to nonhost. We set out to understand the genetic architecture underlying resistance to the biotrophic fungal pathogen Puccinia striiformis f. sp. tritici (wheat stripe rust) in intermediate host and nonhost grass species. In the diploid grass species Hordeum vulgare (barley) and Brachypodium distachyon (purple false brome) we have identified several loci that contribute to limiting the host range of P. striiformis. Mapping of nonhost resistance in both species has found that resistance is conditioned by a few major effect loci. We have fine-mapped six nonhost resistance loci, including Rps6, Rps7, and Rps8 in barley, and Yrr1, Yrr2, and Yrr3 in B. distachyon. Ongoing work in the group includes map-based cloning of these loci and their association with known and novel components of immunity.